Heading here
My First paragraph
My second paragraph
from dsc80_utils import *
Goal: Access information about HDSI faculty members from the HDSI Faculty page.
Let's start by making a GET request to the HDSI Faculty page and see what the resulting HTML looks like.
import requests
fac_response = requests.get('https://datascience.ucsd.edu/faculty/', verify=False)
fac_response
/Users/msgol/ENTER/envs/dsc80/lib/python3.12/site-packages/urllib3/connectionpool.py:1099: InsecureRequestWarning: Unverified HTTPS request is being made to host 'datascience.ucsd.edu'. Adding certificate verification is strongly advised. See: https://urllib3.readthedocs.io/en/latest/advanced-usage.html#tls-warnings warnings.warn(
<Response [200]>
fac_text = fac_response.text
len(fac_text)
257139
print(fac_text[:1000])
<!DOCTYPE html><html lang="en-US"><head><meta charset="UTF-8"><meta name="viewport" content="width=device-width, initial-scale=1.0" /><meta http-equiv="X-UA-Compatible" content="IE=edge"><link rel="profile" href="https://gmpg.org/xfn/11" /><title>Faculty – Halıcıoğlu Data Science Institute – UC San Diego</title> <script defer src="data:text/javascript;base64,CiAgICAgICAgICAgICAgICAgICAgICAgICAgICAvKiBZb3UgY2FuIGFkZCBtb3JlIGNvbmZpZ3VyYXRpb24gb3B0aW9ucyB0byB3ZWJmb250bG9hZGVyIGJ5IHByZXZpb3VzbHkgZGVmaW5pbmcgdGhlIFdlYkZvbnRDb25maWcgd2l0aCB5b3VyIG9wdGlvbnMgKi8KICAgICAgICAgICAgICAgICAgICAgICAgICAgIGlmICggdHlwZW9mIFdlYkZvbnRDb25maWcgPT09ICJ1bmRlZmluZWQiICkgewogICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIFdlYkZvbnRDb25maWcgPSBuZXcgT2JqZWN0KCk7CiAgICAgICAgICAgICAgICAgICAgICAgICAgICB9CiAgICAgICAgICAgICAgICAgICAgICAgICAgICBXZWJGb250Q29uZmlnWydnb29nbGUnXSA9IHtmYW1pbGllczogWydSb2JvdG86NDAwLDUwMCcsICdKb3N0OjQwMCw3MDAnLCAnVGVrbzo3MDAnXX07CgogICAgICAgICAgICAgICAgICAgICAgICAgICAgKGZ1bmN0aW9uKCk
This is not super intuitive to read!
For instance, here's the content of a very basic webpage.
# Mac or Linux:
!cat data/example1.html
# Windows:
# !type data\example1.html
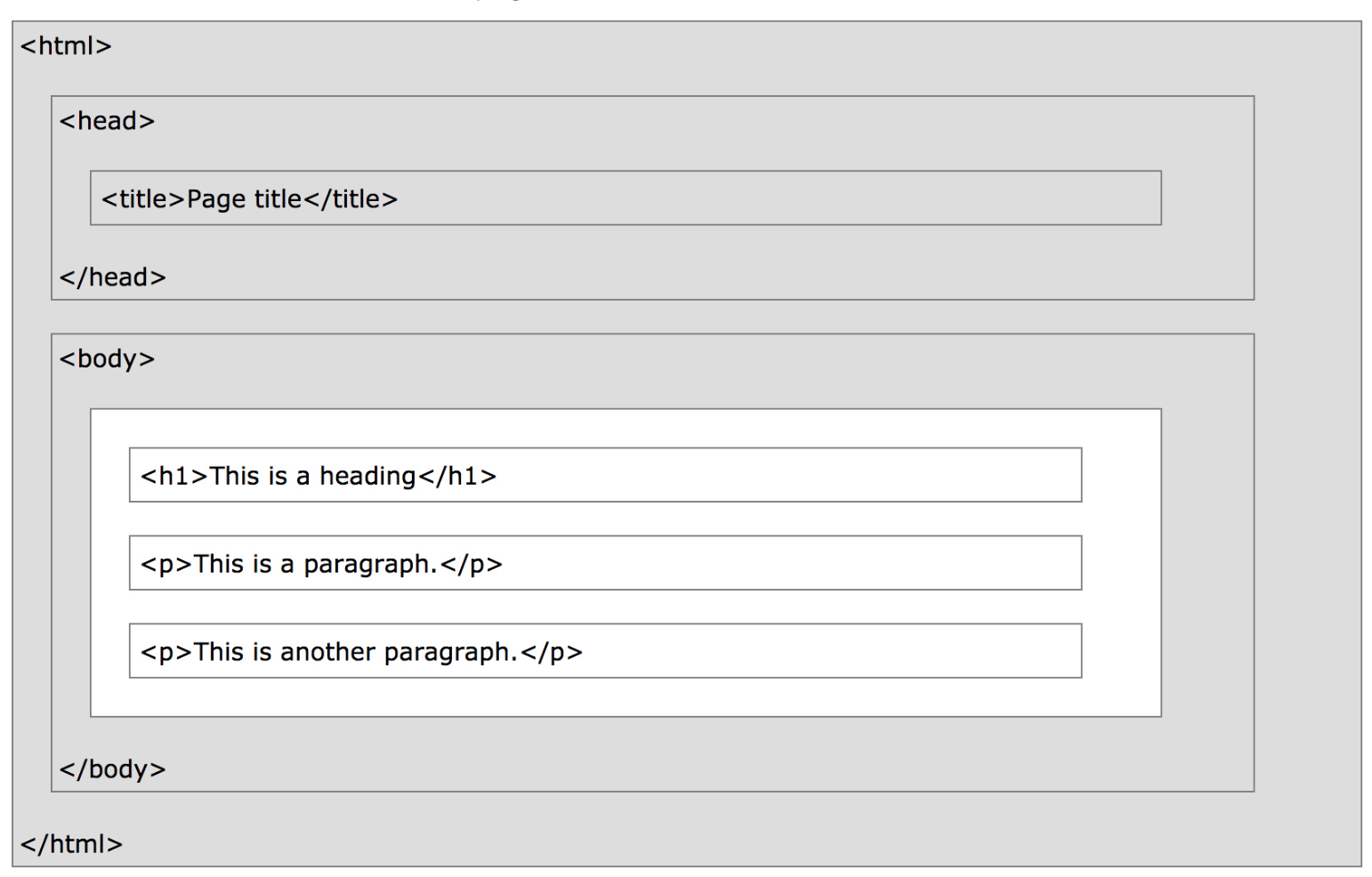
<html>
<head>
<title>Page title</title>
</head>
<body>
<h1>This is a heading</h1>
<p>This is a paragraph.</p>
<p>This is <b>another</b> paragraph.</p>
</body>
</html>
Using IPython.display.HTML, we can render it directly in our notebook.
from IPython.display import HTML
HTML(filename=Path('data') / 'example1.html')
This is a paragraph.
This is another paragraph.
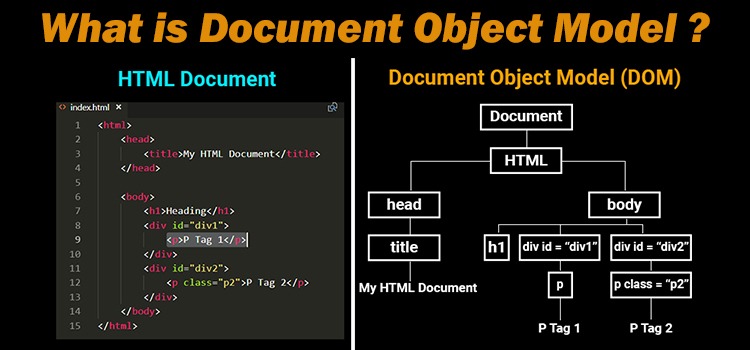
HTML document: The totality of markup that makes up a webpage.
Document Object Model (DOM): The internal representation of an HTML document as a hierarchical tree structure.
HTML element: An object in the DOM, such as a paragraph, header, or title.
HTML tags: Markers that denote the start and end of an element, such as <p> and </p>.
| Element | Description |
|---|---|
<html> |
the document |
<head> |
the header |
<body> |
the body |
<div> |
a logical division of the document |
<span> |
an inline logical division |
<p> |
a paragraph |
<a> |
an anchor (hyperlink) |
<h1>, <h2>, ... |
header(s) |
<img> |
an image |
There are many, many more, but these are by far the most common. See this article for examples.
Tags can have attributes, which further specify how to display information on a webpage.
For instance, <img> tags have src and alt attributes (among others):
<img src="king-selfie.png" alt="A photograph of King Triton." width=500>
Hyperlinks have href attributes:
Click <a href="https://practice.dsc80.com">this link</a> to access past exams.
What do you think this webpage looks like?
# Mac or Linux:
!cat data/example2.html
# Windows:
# !type data\example2.html
<html>
<head>
<title>Example</title>
</head>
<body>
<h1>Foodie Faces</h1>
<img src="../imgs/lunch.jpg" width="200" alt="avocado toast face" />
<img src="../imgs/breakfast.jpg" width="200" alt="Trump oatmeal" />
<p>
These meals were inspired by the book Foodie Faces by Bill Wurtzel and Claire Wurtzel, available on
<a href="https://www.amazon.com/Foodie-Faces-Bill-Wurtzel/dp/0316423548"
>Amazon</a
>.
</p>
<center>
<h3>
Hope this made you smile!
</h3>
</center>
</body>
</html>
<div> tag¶<div style="background-color:lightblue">
<h3>This is a heading</h3>
<p>This is a paragraph.</p>
</div>
The <div> tag defines a division or a "section" of an HTML document.
<div> as a "cell" in a Jupyter Notebook.The <div> element is often used as a container for other HTML elements to style them with CSS or to perform operations involving them using JavaScript.
<div> elements often have attributes, which are important when scraping!
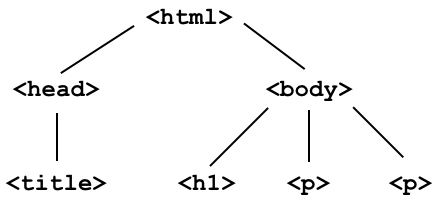
Under the document object model (DOM), HTML documents are trees. In DOM trees, child nodes are ordered.
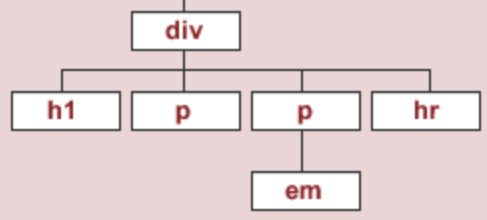
What does the DOM tree look like for this document?
To start, we'll work with the source code for an HTML page with the DOM tree shown below:
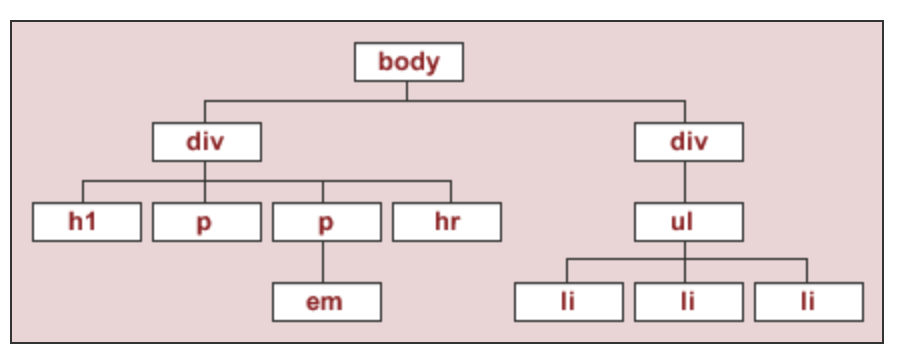
The string html_string contains an HTML "document".
html_string = '''
<html>
<body>
<div id="content">
<h1>Heading here</h1>
<p>My First paragraph</p>
<p>My <em>second</em> paragraph</p>
<hr>
</div>
<div id="nav">
<ul>
<li>item 1</li>
<li>item 2</li>
<li>item 3</li>
</ul>
</div>
</body>
</html>
'''.strip()
HTML(html_string)
My First paragraph
My second paragraph
BeautifulSoup objects¶bs4.BeautifulSoup takes in a string or file-like object representing HTML (markup) and returns a parsed document.
import bs4
bs4.BeautifulSoup?
Init signature: bs4.BeautifulSoup( markup='', features=None, builder=None, parse_only=None, from_encoding=None, exclude_encodings=None, element_classes=None, **kwargs, ) Docstring: A data structure representing a parsed HTML or XML document. Most of the methods you'll call on a BeautifulSoup object are inherited from PageElement or Tag. Internally, this class defines the basic interface called by the tree builders when converting an HTML/XML document into a data structure. The interface abstracts away the differences between parsers. To write a new tree builder, you'll need to understand these methods as a whole. These methods will be called by the BeautifulSoup constructor: * reset() * feed(markup) The tree builder may call these methods from its feed() implementation: * handle_starttag(name, attrs) # See note about return value * handle_endtag(name) * handle_data(data) # Appends to the current data node * endData(containerClass) # Ends the current data node No matter how complicated the underlying parser is, you should be able to build a tree using 'start tag' events, 'end tag' events, 'data' events, and "done with data" events. If you encounter an empty-element tag (aka a self-closing tag, like HTML's <br> tag), call handle_starttag and then handle_endtag. Init docstring: Constructor. :param markup: A string or a file-like object representing markup to be parsed. :param features: Desirable features of the parser to be used. This may be the name of a specific parser ("lxml", "lxml-xml", "html.parser", or "html5lib") or it may be the type of markup to be used ("html", "html5", "xml"). It's recommended that you name a specific parser, so that Beautiful Soup gives you the same results across platforms and virtual environments. :param builder: A TreeBuilder subclass to instantiate (or instance to use) instead of looking one up based on `features`. You only need to use this if you've implemented a custom TreeBuilder. :param parse_only: A SoupStrainer. Only parts of the document matching the SoupStrainer will be considered. This is useful when parsing part of a document that would otherwise be too large to fit into memory. :param from_encoding: A string indicating the encoding of the document to be parsed. Pass this in if Beautiful Soup is guessing wrongly about the document's encoding. :param exclude_encodings: A list of strings indicating encodings known to be wrong. Pass this in if you don't know the document's encoding but you know Beautiful Soup's guess is wrong. :param element_classes: A dictionary mapping BeautifulSoup classes like Tag and NavigableString, to other classes you'd like to be instantiated instead as the parse tree is built. This is useful for subclassing Tag or NavigableString to modify default behavior. :param kwargs: For backwards compatibility purposes, the constructor accepts certain keyword arguments used in Beautiful Soup 3. None of these arguments do anything in Beautiful Soup 4; they will result in a warning and then be ignored. Apart from this, any keyword arguments passed into the BeautifulSoup constructor are propagated to the TreeBuilder constructor. This makes it possible to configure a TreeBuilder by passing in arguments, not just by saying which one to use. File: ~/ENTER/envs/dsc80/lib/python3.12/site-packages/bs4/__init__.py Type: type Subclasses: BeautifulStoneSoup
Normally, we pass the result of a GET request to bs4.BeautifulSoup, but here we will pass our hand-crafted html_string.
soup = bs4.BeautifulSoup(html_string)
soup
<html> <body> <div id="content"> <h1>Heading here</h1> <p>My First paragraph</p> <p>My <em>second</em> paragraph</p> <hr/> </div> <div id="nav"> <ul> <li>item 1</li> <li>item 2</li> <li>item 3</li> </ul> </div> </body> </html>
type(soup)
bs4.BeautifulSoup
BeautifulSoup objects have several useful attributes, e.g. text:
print(soup.text)
Heading here My First paragraph My second paragraph item 1 item 2 item 3
descendants¶The descendants attribute traverses a BeautifulSoup tree using depth-first traversal.
Why depth-first? Elements closer to one another on a page are more likely to be related than elements further away.

soup.descendants
<generator object Tag.descendants at 0x1067297d0>
for child in soup.descendants:
# print(child) # What would happen if we ran this instead?
if not isinstance(child, str):
print(child.name)
html body div h1 p p em hr div ul li li li
Practically speaking, you will not use the descendants attribute (or the related children attribute) directly very often. Instead, you will use the following methods:
soup.find(tag), which finds the first instance of a tag (the first one on the page, i.e. the first one that DFS sees).soup.find(name=None, attrs={}, recursive=True, text=None, **kwargs).soup.find_all(tag) will find all instances of a tag.find finds tags!
soup.find('div')
<div id="content"> <h1>Heading here</h1> <p>My First paragraph</p> <p>My <em>second</em> paragraph</p> <hr/> </div>
Let's try to find the <div> element that has an id attribute equal to 'nav'.
soup.find('div', attrs={'id': 'nav'})
<div id="nav"> <ul> <li>item 1</li> <li>item 2</li> <li>item 3</li> </ul> </div>
find will return the first occurrence of a tag, regardless of its depth in the tree.
# Three li children - we get the first one.
soup.find('li')
<li>item 1</li>
find_all¶find_all returns a list of all matches.
soup.find_all('div')
[<div id="content"> <h1>Heading here</h1> <p>My First paragraph</p> <p>My <em>second</em> paragraph</p> <hr/> </div>, <div id="nav"> <ul> <li>item 1</li> <li>item 2</li> <li>item 3</li> </ul> </div>]
soup.find_all('li')
[<li>item 1</li>, <li>item 2</li>, <li>item 3</li>]
[x.text for x in soup.find_all('li')]
['item 1', 'item 2', 'item 3']
text attribute of a tag element gets the text between the opening and closing tags.attrs attribute of a tag element lists all of its attributes.get method of a tag element gets the value of an attribute.soup.find('p')
<p>My First paragraph</p>
soup.find('p').text
'My First paragraph'
soup.find('div')
<div id="content"> <h1>Heading here</h1> <p>My First paragraph</p> <p>My <em>second</em> paragraph</p> <hr/> </div>
soup.find('div').text
'\nHeading here\nMy First paragraph\nMy second paragraph\n\n'
soup.find('div').attrs
{'id': 'content'}
soup.find('div').get('id')
'content'
The get method must be called directly on the node that contains the attribute you're looking for.
soup
<html> <body> <div id="content"> <h1>Heading here</h1> <p>My First paragraph</p> <p>My <em>second</em> paragraph</p> <hr/> </div> <div id="nav"> <ul> <li>item 1</li> <li>item 2</li> <li>item 3</li> </ul> </div> </body> </html>
# While there are multiple 'id' attributes, none of them are in the <html> tag at the top.
soup.get('id')
soup.find('div').get('id')
'content'
Consider the following HTML document.
<html>
<head>
<title>3*Global-2026-02-04</title>
</head>
<body>
<h1>Spotify Top 3 - Global</h1>
<table>
<tr class='heading'>
<th>Rank</th>
<th>Artist</th>
<th>Song</th>
</tr>
<tr class=1>
<td>1</td>
<td>Djo</td>
<td><a href='https://open.spotify.com/track/3qhlB30KknSejmIvZZLjOD'>End of Beginning</a></td>
</tr>
<tr class=2>
<td>2</td>
<td>Harry Styles</td>
<td><a href='https://open.spotify.com/track/45Z3m6yazmAi4jZuW0tzW0'>Aperture</a></td>
</tr>
<tr class=3>
<td>3</td>
<td>Taylor Swift</td>
<td><a href='https://open.spotify.com/track/53iuhJlwXhSER5J2IYYv1W'>The Fate of Ophelia</a></td>
</tr>
</table>
</body>
</html>
Part 1: How many leaf nodes are there in the DOM tree of this document — that is, how many nodes have no children?
# Create a BeautifulSoup object from the Spotify data.
soup = bs4.BeautifulSoup((Path('data') / 'example3.html').read_text())
Part 2: What does the following line of code evaluate to?
len(soup.find_all("td"))
Part 3: What does the following line of code evaluate to?
soup.find("a").get("href")
The website quotes.toscrape.com looks like this:
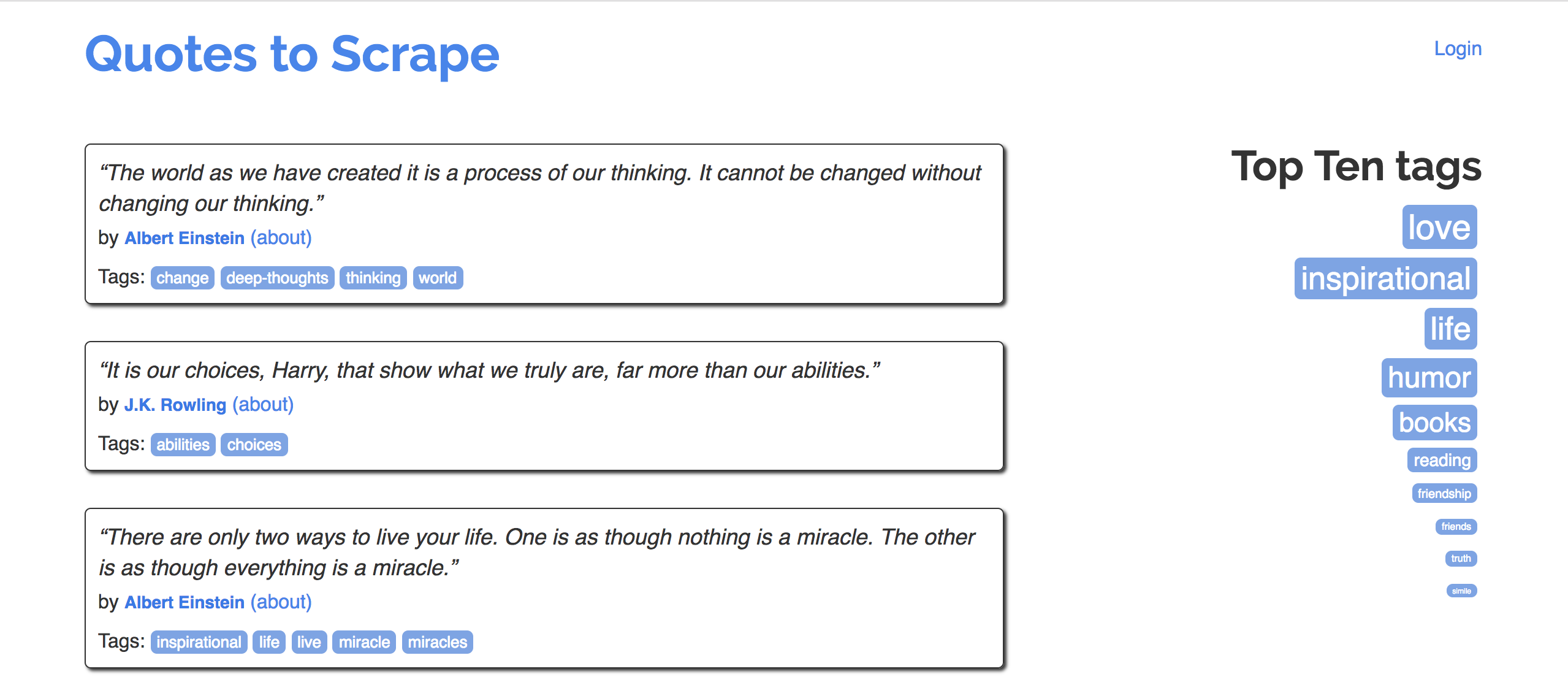
Goal: Extract quotes (and relevant metadata) into a DataFrame.
Specifically, let's try to make a DataFrame that looks like the one below:
| quote | author | author_url | tags | |
|---|---|---|---|---|
| 0 | “The world as we have created it is a process of our thinking. It cannot be changed without changing our thinking.” | Albert Einstein | https://quotes.toscrape.com/author/Albert-Einstein | change,deep-thoughts,thinking,world |
| 1 | “It is our choices, Harry, that show what we truly are, far more than our abilities.” | J.K. Rowling | https://quotes.toscrape.com/author/J-K-Rowling | abilities,choices |
| 2 | “There are only two ways to live your life. One is as though nothing is a miracle. The other is as though everything is a miracle.” | Albert Einstein | https://quotes.toscrape.com/author/Albert-Einstein | inspirational,life,live,miracle,miracles |
Eventually, we will create a single function – make_quote_df – which takes in an integer n and returns a DataFrame with the quotes on the first n pages of quotes.toscrape.com.
To do this, we will define several helper functions:
download_page(i), which downloads a single page (page i) and returns a BeautifulSoup object of the response.
process_quote(div), which takes in a <div> tree corresponding to a single quote and returns a dictionary containing all of the relevant information for that quote.
process_page(divs), which takes in a list of <div> trees corresponding to a single page and returns a DataFrame containing all of the relevant information for all quotes on that page.
Key principle: some of our helper functions will make requests, and others will parse, but none will do both!
def download_page(i):
req = requests.get(f"http://quotes.toscrape.com/page/{i}/")
soup = bs4.BeautifulSoup(req.text)
return soup
In make_quote_df, we will call download_page repeatedly – once for i=1, once for i=2, ..., i=n. For now, we will work with just page 1.
soup = download_page(1)
soup
<!DOCTYPE html>
<html lang="en">
<head>
<meta charset="utf-8"/>
<title>Quotes to Scrape</title>
<link href="/static/bootstrap.min.css" rel="stylesheet"/>
<link href="/static/main.css" rel="stylesheet"/>
</head>
<body>
<div class="container">
<div class="row header-box">
<div class="col-md-8">
<h1>
<a href="/" style="text-decoration: none">Quotes to Scrape</a>
</h1>
</div>
<div class="col-md-4">
<p>
<a href="/login">Login</a>
</p>
</div>
</div>
<div class="row">
<div class="col-md-8">
<div class="quote" itemscope="" itemtype="http://schema.org/CreativeWork">
<span class="text" itemprop="text">“The world as we have created it is a process of our thinking. It cannot be changed without changing our thinking.”</span>
<span>by <small class="author" itemprop="author">Albert Einstein</small>
<a href="/author/Albert-Einstein">(about)</a>
</span>
<div class="tags">
Tags:
<meta class="keywords" content="change,deep-thoughts,thinking,world" itemprop="keywords"/>
<a class="tag" href="/tag/change/page/1/">change</a>
<a class="tag" href="/tag/deep-thoughts/page/1/">deep-thoughts</a>
<a class="tag" href="/tag/thinking/page/1/">thinking</a>
<a class="tag" href="/tag/world/page/1/">world</a>
</div>
</div>
<div class="quote" itemscope="" itemtype="http://schema.org/CreativeWork">
<span class="text" itemprop="text">“It is our choices, Harry, that show what we truly are, far more than our abilities.”</span>
<span>by <small class="author" itemprop="author">J.K. Rowling</small>
<a href="/author/J-K-Rowling">(about)</a>
</span>
<div class="tags">
Tags:
<meta class="keywords" content="abilities,choices" itemprop="keywords"/>
<a class="tag" href="/tag/abilities/page/1/">abilities</a>
<a class="tag" href="/tag/choices/page/1/">choices</a>
</div>
</div>
<div class="quote" itemscope="" itemtype="http://schema.org/CreativeWork">
<span class="text" itemprop="text">“There are only two ways to live your life. One is as though nothing is a miracle. The other is as though everything is a miracle.”</span>
<span>by <small class="author" itemprop="author">Albert Einstein</small>
<a href="/author/Albert-Einstein">(about)</a>
</span>
<div class="tags">
Tags:
<meta class="keywords" content="inspirational,life,live,miracle,miracles" itemprop="keywords"/>
<a class="tag" href="/tag/inspirational/page/1/">inspirational</a>
<a class="tag" href="/tag/life/page/1/">life</a>
<a class="tag" href="/tag/live/page/1/">live</a>
<a class="tag" href="/tag/miracle/page/1/">miracle</a>
<a class="tag" href="/tag/miracles/page/1/">miracles</a>
</div>
</div>
<div class="quote" itemscope="" itemtype="http://schema.org/CreativeWork">
<span class="text" itemprop="text">“The person, be it gentleman or lady, who has not pleasure in a good novel, must be intolerably stupid.”</span>
<span>by <small class="author" itemprop="author">Jane Austen</small>
<a href="/author/Jane-Austen">(about)</a>
</span>
<div class="tags">
Tags:
<meta class="keywords" content="aliteracy,books,classic,humor" itemprop="keywords"/>
<a class="tag" href="/tag/aliteracy/page/1/">aliteracy</a>
<a class="tag" href="/tag/books/page/1/">books</a>
<a class="tag" href="/tag/classic/page/1/">classic</a>
<a class="tag" href="/tag/humor/page/1/">humor</a>
</div>
</div>
<div class="quote" itemscope="" itemtype="http://schema.org/CreativeWork">
<span class="text" itemprop="text">“Imperfection is beauty, madness is genius and it's better to be absolutely ridiculous than absolutely boring.”</span>
<span>by <small class="author" itemprop="author">Marilyn Monroe</small>
<a href="/author/Marilyn-Monroe">(about)</a>
</span>
<div class="tags">
Tags:
<meta class="keywords" content="be-yourself,inspirational" itemprop="keywords"/>
<a class="tag" href="/tag/be-yourself/page/1/">be-yourself</a>
<a class="tag" href="/tag/inspirational/page/1/">inspirational</a>
</div>
</div>
<div class="quote" itemscope="" itemtype="http://schema.org/CreativeWork">
<span class="text" itemprop="text">“Try not to become a man of success. Rather become a man of value.”</span>
<span>by <small class="author" itemprop="author">Albert Einstein</small>
<a href="/author/Albert-Einstein">(about)</a>
</span>
<div class="tags">
Tags:
<meta class="keywords" content="adulthood,success,value" itemprop="keywords"/>
<a class="tag" href="/tag/adulthood/page/1/">adulthood</a>
<a class="tag" href="/tag/success/page/1/">success</a>
<a class="tag" href="/tag/value/page/1/">value</a>
</div>
</div>
<div class="quote" itemscope="" itemtype="http://schema.org/CreativeWork">
<span class="text" itemprop="text">“It is better to be hated for what you are than to be loved for what you are not.”</span>
<span>by <small class="author" itemprop="author">André Gide</small>
<a href="/author/Andre-Gide">(about)</a>
</span>
<div class="tags">
Tags:
<meta class="keywords" content="life,love" itemprop="keywords"/>
<a class="tag" href="/tag/life/page/1/">life</a>
<a class="tag" href="/tag/love/page/1/">love</a>
</div>
</div>
<div class="quote" itemscope="" itemtype="http://schema.org/CreativeWork">
<span class="text" itemprop="text">“I have not failed. I've just found 10,000 ways that won't work.”</span>
<span>by <small class="author" itemprop="author">Thomas A. Edison</small>
<a href="/author/Thomas-A-Edison">(about)</a>
</span>
<div class="tags">
Tags:
<meta class="keywords" content="edison,failure,inspirational,paraphrased" itemprop="keywords"/>
<a class="tag" href="/tag/edison/page/1/">edison</a>
<a class="tag" href="/tag/failure/page/1/">failure</a>
<a class="tag" href="/tag/inspirational/page/1/">inspirational</a>
<a class="tag" href="/tag/paraphrased/page/1/">paraphrased</a>
</div>
</div>
<div class="quote" itemscope="" itemtype="http://schema.org/CreativeWork">
<span class="text" itemprop="text">“A woman is like a tea bag; you never know how strong it is until it's in hot water.”</span>
<span>by <small class="author" itemprop="author">Eleanor Roosevelt</small>
<a href="/author/Eleanor-Roosevelt">(about)</a>
</span>
<div class="tags">
Tags:
<meta class="keywords" content="misattributed-eleanor-roosevelt" itemprop="keywords"/>
<a class="tag" href="/tag/misattributed-eleanor-roosevelt/page/1/">misattributed-eleanor-roosevelt</a>
</div>
</div>
<div class="quote" itemscope="" itemtype="http://schema.org/CreativeWork">
<span class="text" itemprop="text">“A day without sunshine is like, you know, night.”</span>
<span>by <small class="author" itemprop="author">Steve Martin</small>
<a href="/author/Steve-Martin">(about)</a>
</span>
<div class="tags">
Tags:
<meta class="keywords" content="humor,obvious,simile" itemprop="keywords"/>
<a class="tag" href="/tag/humor/page/1/">humor</a>
<a class="tag" href="/tag/obvious/page/1/">obvious</a>
<a class="tag" href="/tag/simile/page/1/">simile</a>
</div>
</div>
<nav>
<ul class="pager">
<li class="next">
<a href="/page/2/">Next <span aria-hidden="true">→</span></a>
</li>
</ul>
</nav>
</div>
<div class="col-md-4 tags-box">
<h2>Top Ten tags</h2>
<span class="tag-item">
<a class="tag" href="/tag/love/" style="font-size: 28px">love</a>
</span>
<span class="tag-item">
<a class="tag" href="/tag/inspirational/" style="font-size: 26px">inspirational</a>
</span>
<span class="tag-item">
<a class="tag" href="/tag/life/" style="font-size: 26px">life</a>
</span>
<span class="tag-item">
<a class="tag" href="/tag/humor/" style="font-size: 24px">humor</a>
</span>
<span class="tag-item">
<a class="tag" href="/tag/books/" style="font-size: 22px">books</a>
</span>
<span class="tag-item">
<a class="tag" href="/tag/reading/" style="font-size: 14px">reading</a>
</span>
<span class="tag-item">
<a class="tag" href="/tag/friendship/" style="font-size: 10px">friendship</a>
</span>
<span class="tag-item">
<a class="tag" href="/tag/friends/" style="font-size: 8px">friends</a>
</span>
<span class="tag-item">
<a class="tag" href="/tag/truth/" style="font-size: 8px">truth</a>
</span>
<span class="tag-item">
<a class="tag" href="/tag/simile/" style="font-size: 6px">simile</a>
</span>
</div>
</div>
</div>
<footer class="footer">
<div class="container">
<p class="text-muted">
Quotes by: <a href="https://www.goodreads.com/quotes">GoodReads.com</a>
</p>
<p class="copyright">
Made with <span class="zyte">❤</span> by <a class="zyte" href="https://www.zyte.com">Zyte</a>
</p>
</div>
</footer>
</body>
</html>
Let's look at the page's source code (right click the page and click "Inspect" in Chrome) to find where the quotes in the page are located.
divs = soup.find_all('div',attrs={'class','quote'})
#Now write the equivalent one below with special class_ keyword attribute
divs
[<div class="quote" itemscope="" itemtype="http://schema.org/CreativeWork">
<span class="text" itemprop="text">“The world as we have created it is a process of our thinking. It cannot be changed without changing our thinking.”</span>
<span>by <small class="author" itemprop="author">Albert Einstein</small>
<a href="/author/Albert-Einstein">(about)</a>
</span>
<div class="tags">
Tags:
<meta class="keywords" content="change,deep-thoughts,thinking,world" itemprop="keywords"/>
<a class="tag" href="/tag/change/page/1/">change</a>
<a class="tag" href="/tag/deep-thoughts/page/1/">deep-thoughts</a>
<a class="tag" href="/tag/thinking/page/1/">thinking</a>
<a class="tag" href="/tag/world/page/1/">world</a>
</div>
</div>,
<div class="quote" itemscope="" itemtype="http://schema.org/CreativeWork">
<span class="text" itemprop="text">“It is our choices, Harry, that show what we truly are, far more than our abilities.”</span>
<span>by <small class="author" itemprop="author">J.K. Rowling</small>
<a href="/author/J-K-Rowling">(about)</a>
</span>
<div class="tags">
Tags:
<meta class="keywords" content="abilities,choices" itemprop="keywords"/>
<a class="tag" href="/tag/abilities/page/1/">abilities</a>
<a class="tag" href="/tag/choices/page/1/">choices</a>
</div>
</div>,
<div class="quote" itemscope="" itemtype="http://schema.org/CreativeWork">
<span class="text" itemprop="text">“There are only two ways to live your life. One is as though nothing is a miracle. The other is as though everything is a miracle.”</span>
<span>by <small class="author" itemprop="author">Albert Einstein</small>
<a href="/author/Albert-Einstein">(about)</a>
</span>
<div class="tags">
Tags:
<meta class="keywords" content="inspirational,life,live,miracle,miracles" itemprop="keywords"/>
<a class="tag" href="/tag/inspirational/page/1/">inspirational</a>
<a class="tag" href="/tag/life/page/1/">life</a>
<a class="tag" href="/tag/live/page/1/">live</a>
<a class="tag" href="/tag/miracle/page/1/">miracle</a>
<a class="tag" href="/tag/miracles/page/1/">miracles</a>
</div>
</div>,
<div class="quote" itemscope="" itemtype="http://schema.org/CreativeWork">
<span class="text" itemprop="text">“The person, be it gentleman or lady, who has not pleasure in a good novel, must be intolerably stupid.”</span>
<span>by <small class="author" itemprop="author">Jane Austen</small>
<a href="/author/Jane-Austen">(about)</a>
</span>
<div class="tags">
Tags:
<meta class="keywords" content="aliteracy,books,classic,humor" itemprop="keywords"/>
<a class="tag" href="/tag/aliteracy/page/1/">aliteracy</a>
<a class="tag" href="/tag/books/page/1/">books</a>
<a class="tag" href="/tag/classic/page/1/">classic</a>
<a class="tag" href="/tag/humor/page/1/">humor</a>
</div>
</div>,
<div class="quote" itemscope="" itemtype="http://schema.org/CreativeWork">
<span class="text" itemprop="text">“Imperfection is beauty, madness is genius and it's better to be absolutely ridiculous than absolutely boring.”</span>
<span>by <small class="author" itemprop="author">Marilyn Monroe</small>
<a href="/author/Marilyn-Monroe">(about)</a>
</span>
<div class="tags">
Tags:
<meta class="keywords" content="be-yourself,inspirational" itemprop="keywords"/>
<a class="tag" href="/tag/be-yourself/page/1/">be-yourself</a>
<a class="tag" href="/tag/inspirational/page/1/">inspirational</a>
</div>
</div>,
<div class="quote" itemscope="" itemtype="http://schema.org/CreativeWork">
<span class="text" itemprop="text">“Try not to become a man of success. Rather become a man of value.”</span>
<span>by <small class="author" itemprop="author">Albert Einstein</small>
<a href="/author/Albert-Einstein">(about)</a>
</span>
<div class="tags">
Tags:
<meta class="keywords" content="adulthood,success,value" itemprop="keywords"/>
<a class="tag" href="/tag/adulthood/page/1/">adulthood</a>
<a class="tag" href="/tag/success/page/1/">success</a>
<a class="tag" href="/tag/value/page/1/">value</a>
</div>
</div>,
<div class="quote" itemscope="" itemtype="http://schema.org/CreativeWork">
<span class="text" itemprop="text">“It is better to be hated for what you are than to be loved for what you are not.”</span>
<span>by <small class="author" itemprop="author">André Gide</small>
<a href="/author/Andre-Gide">(about)</a>
</span>
<div class="tags">
Tags:
<meta class="keywords" content="life,love" itemprop="keywords"/>
<a class="tag" href="/tag/life/page/1/">life</a>
<a class="tag" href="/tag/love/page/1/">love</a>
</div>
</div>,
<div class="quote" itemscope="" itemtype="http://schema.org/CreativeWork">
<span class="text" itemprop="text">“I have not failed. I've just found 10,000 ways that won't work.”</span>
<span>by <small class="author" itemprop="author">Thomas A. Edison</small>
<a href="/author/Thomas-A-Edison">(about)</a>
</span>
<div class="tags">
Tags:
<meta class="keywords" content="edison,failure,inspirational,paraphrased" itemprop="keywords"/>
<a class="tag" href="/tag/edison/page/1/">edison</a>
<a class="tag" href="/tag/failure/page/1/">failure</a>
<a class="tag" href="/tag/inspirational/page/1/">inspirational</a>
<a class="tag" href="/tag/paraphrased/page/1/">paraphrased</a>
</div>
</div>,
<div class="quote" itemscope="" itemtype="http://schema.org/CreativeWork">
<span class="text" itemprop="text">“A woman is like a tea bag; you never know how strong it is until it's in hot water.”</span>
<span>by <small class="author" itemprop="author">Eleanor Roosevelt</small>
<a href="/author/Eleanor-Roosevelt">(about)</a>
</span>
<div class="tags">
Tags:
<meta class="keywords" content="misattributed-eleanor-roosevelt" itemprop="keywords"/>
<a class="tag" href="/tag/misattributed-eleanor-roosevelt/page/1/">misattributed-eleanor-roosevelt</a>
</div>
</div>,
<div class="quote" itemscope="" itemtype="http://schema.org/CreativeWork">
<span class="text" itemprop="text">“A day without sunshine is like, you know, night.”</span>
<span>by <small class="author" itemprop="author">Steve Martin</small>
<a href="/author/Steve-Martin">(about)</a>
</span>
<div class="tags">
Tags:
<meta class="keywords" content="humor,obvious,simile" itemprop="keywords"/>
<a class="tag" href="/tag/humor/page/1/">humor</a>
<a class="tag" href="/tag/obvious/page/1/">obvious</a>
<a class="tag" href="/tag/simile/page/1/">simile</a>
</div>
</div>]
From each <div>, we can extract the quote, author name, author's URL, and tags.
divs[0]
<div class="quote" itemscope="" itemtype="http://schema.org/CreativeWork">
<span class="text" itemprop="text">“The world as we have created it is a process of our thinking. It cannot be changed without changing our thinking.”</span>
<span>by <small class="author" itemprop="author">Albert Einstein</small>
<a href="/author/Albert-Einstein">(about)</a>
</span>
<div class="tags">
Tags:
<meta class="keywords" content="change,deep-thoughts,thinking,world" itemprop="keywords"/>
<a class="tag" href="/tag/change/page/1/">change</a>
<a class="tag" href="/tag/deep-thoughts/page/1/">deep-thoughts</a>
<a class="tag" href="/tag/thinking/page/1/">thinking</a>
<a class="tag" href="/tag/world/page/1/">world</a>
</div>
</div>
# The quote.
divs[0].find('span', attrs={'class':'text'}).text
'“The world as we have created it is a process of our thinking. It cannot be changed without changing our thinking.”'
# The author.
divs[0].find('small', class_='author').text
'Albert Einstein'
# The URL for the author.
"quotes.toscrape.com" + divs[0].find('a').get('href')
'quotes.toscrape.com/author/Albert-Einstein'
# The quote's tags.
divs[0].find('meta').get('content')
'change,deep-thoughts,thinking,world'
Let's implement our next function, process_quote, which takes in a <div> corresponding to a single quote and returns a dictionary containing the quote's information.
Why use a dictionary? Passing pd.DataFrame a list of dictionaries is an easy way to create a DataFrame.
def process_quote(div):
quote = div.find('span', class_='text').text
author = div.find('small', class_='author').text
author_url = 'https://quotes.toscrape.com' + div.find('a').get('href')
tags = div.find('meta', class_='keywords').get('content')
return {'quote': quote, 'author': author, 'author_url': author_url, 'tags': tags}
process_quote(divs[-1])
{'quote': '“A day without sunshine is like, you know, night.”',
'author': 'Steve Martin',
'author_url': 'https://quotes.toscrape.com/author/Steve-Martin',
'tags': 'humor,obvious,simile'}
Our last helper function will take in a list of <div>s, call process_quote on each <div> in the list, and return a DataFrame.
def process_page(divs):
return pd.DataFrame([process_quote(div) for div in divs])
process_page(divs)
| quote | author | author_url | tags | |
|---|---|---|---|---|
| 0 | “The world as we have created it is a process ... | Albert Einstein | https://quotes.toscrape.com/author/Albert-Eins... | change,deep-thoughts,thinking,world |
| 1 | “It is our choices, Harry, that show what we t... | J.K. Rowling | https://quotes.toscrape.com/author/J-K-Rowling | abilities,choices |
| 2 | “There are only two ways to live your life. On... | Albert Einstein | https://quotes.toscrape.com/author/Albert-Eins... | inspirational,life,live,miracle,miracles |
| ... | ... | ... | ... | ... |
| 7 | “I have not failed. I've just found 10,000 way... | Thomas A. Edison | https://quotes.toscrape.com/author/Thomas-A-Ed... | edison,failure,inspirational,paraphrased |
| 8 | “A woman is like a tea bag; you never know how... | Eleanor Roosevelt | https://quotes.toscrape.com/author/Eleanor-Roo... | misattributed-eleanor-roosevelt |
| 9 | “A day without sunshine is like, you know, nig... | Steve Martin | https://quotes.toscrape.com/author/Steve-Martin | humor,obvious,simile |
10 rows × 4 columns
def make_quote_df(n):
'''Returns a DataFrame containing the quotes on the first n pages of https://quotes.toscrape.com/.'''
dfs = []
for i in range(1, n+1):
# Download page n and create a BeautifulSoup object.
soup = download_page(i)
# Create a DataFrame of the information on that page.
divs = soup.find_all('div', class_='quote')
df = process_page(divs)
# Append DataFrame to dfs.
dfs.append(df)
# Stitch all DataFrames together.
return pd.concat(dfs).reset_index(drop=True)
quotes = make_quote_df(3)
quotes.head()
| quote | author | author_url | tags | |
|---|---|---|---|---|
| 0 | “The world as we have created it is a process ... | Albert Einstein | https://quotes.toscrape.com/author/Albert-Eins... | change,deep-thoughts,thinking,world |
| 1 | “It is our choices, Harry, that show what we t... | J.K. Rowling | https://quotes.toscrape.com/author/J-K-Rowling | abilities,choices |
| 2 | “There are only two ways to live your life. On... | Albert Einstein | https://quotes.toscrape.com/author/Albert-Eins... | inspirational,life,live,miracle,miracles |
| 3 | “The person, be it gentleman or lady, who has ... | Jane Austen | https://quotes.toscrape.com/author/Jane-Austen | aliteracy,books,classic,humor |
| 4 | “Imperfection is beauty, madness is genius and... | Marilyn Monroe | https://quotes.toscrape.com/author/Marilyn-Monroe | be-yourself,inspirational |
quotes[quotes['author'] == 'Dr. Seuss']
| quote | author | author_url | tags | |
|---|---|---|---|---|
| 14 | “I like nonsense, it wakes up the brain cells.... | Dr. Seuss | https://quotes.toscrape.com/author/Dr-Seuss | fantasy |
| 25 | “Today you are You, that is truer than true. T... | Dr. Seuss | https://quotes.toscrape.com/author/Dr-Seuss | comedy,life,yourself |
The elements in the 'tags' column are all strings, but they look like lists. This is not ideal, as we will see shortly.
Let's try to extract a list of HDSI Faculty from datascience.ucsd.edu/faculty.
fac_response = requests.get('https://datascience.ucsd.edu/faculty/', verify=False)
fac_response
/Users/msgol/ENTER/envs/dsc80/lib/python3.12/site-packages/urllib3/connectionpool.py:1099: InsecureRequestWarning: Unverified HTTPS request is being made to host 'datascience.ucsd.edu'. Adding certificate verification is strongly advised. See: https://urllib3.readthedocs.io/en/latest/advanced-usage.html#tls-warnings warnings.warn(
<Response [200]>
soup = bs4.BeautifulSoup(fac_response.text)
It's not easy identifying which <div>s we want. The Inspect tool makes this easier, but it's good to verify that find_all is finding the right number of elements.
divs = soup.find_all('div', class_='vc_grid-item')
len(divs)
66
Within here, we need to extract each faculty member's name. It seems like names are stored as text within the <h4> tag.
divs[0]
<div class="vc_grid-item vc_clearfix col_1-5 vc_grid-item-zone-c-bottom vc_visible-item vc_grid-term-faculty-fellows vc_grid-term-council vc_grid-term-faculty"> <a class="anchor-link" id="ilkay-altintas" name="ilkay-altintas"></a><div class="vc_grid-item-mini vc_clearfix"><div class="vc_gitem-animated-block"><div class="vc_gitem-zone vc_gitem-zone-a vc-gitem-zone-height-mode-auto vc-gitem-zone-height-mode-auto-1-1" style="background-image: url(https://datascience.ucsd.edu/wp-content/uploads/2022/10/ilkayaltintas_headshot.jpg) !important;"><a class="vc_gitem-link vc-zone-link" href="https://datascience.ucsd.edu/people/ilkay-altintas/"></a><img alt="Ilkay Altintas" class="vc_gitem-zone-img" decoding="async" src="https://datascience.ucsd.edu/wp-content/uploads/2022/10/ilkayaltintas_headshot.jpg"/><div class="vc_gitem-zone-mini"></div></div></div><div class="vc_gitem-zone vc_gitem-zone-c"><div class="vc_gitem-zone-mini"><div class="vc_gitem_row vc_row vc_gitem-row-position-top"><div class="vc_col-sm-12 vc_gitem-col vc_gitem-col-align-"><div class="vc_custom_heading vc_gitem-post-data vc_gitem-post-data-source-post_title"><h4 style="text-align: left"><a href="https://datascience.ucsd.edu/people/ilkay-altintas/">Ilkay Altintas</a></h4></div><div class="vc_gitem-align-left fields"><div class="field pendari_people_title">SDSC Chief Data Science Officer & HDSI Founding Faculty Fellow</div></div><div class="excerpt"></div><div class="terms"> Faculty Fellows Council Faculty</div></div></div></div></div></div></div>
divs[0].find('h4').text
'Ilkay Altintas'
We can also extract job titles:
divs[0].find(class_='pendari_people_title').text
'SDSC Chief Data Science Officer & HDSI Founding Faculty Fellow'
Let's create a DataFrame consisting of names and job titles for each faculty member.
names = [div.find('h4').text for div in divs]
names[:10]
['Ilkay Altintas', 'Tiffany Amariuta', 'Mikio Aoi', 'Ery Arias-Castro', 'Vineet Bafna', 'Mikhail Belkin', 'Umesh Bellur', 'Peter Chi', 'Henrik Christensen', 'Alex Cloninger']
titles = [div.find(class_='pendari_people_title').text for div in divs]
titles[:10]
['SDSC Chief Data Science Officer & HDSI Founding Faculty Fellow', 'Assistant Professor', 'Assistant Professor', 'Professor', 'Professor', 'Professor', 'Visiting Professor, UCSD HDSI', 'Associate Teaching Professor', 'Distinguished Scientist, Professor', 'Professor']
faculty = pd.DataFrame({
'name': names,
'title': titles,
})
faculty.head()
| name | title | |
|---|---|---|
| 0 | Ilkay Altintas | SDSC Chief Data Science Officer & HDSI Foundin... |
| 1 | Tiffany Amariuta | Assistant Professor |
| 2 | Mikio Aoi | Assistant Professor |
| 3 | Ery Arias-Castro | Professor |
| 4 | Vineet Bafna | Professor |
Now we have a DataFrame!
faculty[faculty['title'].str.contains('Teaching') | faculty['title'].str.contains('Lecturer')]
| name | title | |
|---|---|---|
| 7 | Peter Chi | Associate Teaching Professor |
| 14 | Justin Eldridge | Associate Teaching Professor |
| 15 | Shannon Ellis | Associate Teaching Professor |
| ... | ... | ... |
| 40 | Giorgio Quer | Lecturer |
| 48 | Jack Silberman | Lecturer |
| 52 | Janine Tiefenbruck | Lecturer |
9 rows × 2 columns
What if we want to get faculty members' pictures?
divs[0]
<div class="vc_grid-item vc_clearfix col_1-5 vc_grid-item-zone-c-bottom vc_visible-item vc_grid-term-faculty-fellows vc_grid-term-council vc_grid-term-faculty"> <a class="anchor-link" id="ilkay-altintas" name="ilkay-altintas"></a><div class="vc_grid-item-mini vc_clearfix"><div class="vc_gitem-animated-block"><div class="vc_gitem-zone vc_gitem-zone-a vc-gitem-zone-height-mode-auto vc-gitem-zone-height-mode-auto-1-1" style="background-image: url(https://datascience.ucsd.edu/wp-content/uploads/2022/10/ilkayaltintas_headshot.jpg) !important;"><a class="vc_gitem-link vc-zone-link" href="https://datascience.ucsd.edu/people/ilkay-altintas/"></a><img alt="Ilkay Altintas" class="vc_gitem-zone-img" decoding="async" src="https://datascience.ucsd.edu/wp-content/uploads/2022/10/ilkayaltintas_headshot.jpg"/><div class="vc_gitem-zone-mini"></div></div></div><div class="vc_gitem-zone vc_gitem-zone-c"><div class="vc_gitem-zone-mini"><div class="vc_gitem_row vc_row vc_gitem-row-position-top"><div class="vc_col-sm-12 vc_gitem-col vc_gitem-col-align-"><div class="vc_custom_heading vc_gitem-post-data vc_gitem-post-data-source-post_title"><h4 style="text-align: left"><a href="https://datascience.ucsd.edu/people/ilkay-altintas/">Ilkay Altintas</a></h4></div><div class="vc_gitem-align-left fields"><div class="field pendari_people_title">SDSC Chief Data Science Officer & HDSI Founding Faculty Fellow</div></div><div class="excerpt"></div><div class="terms"> Faculty Fellows Council Faculty</div></div></div></div></div></div></div>
from IPython.display import Image, display
def show_picture(name):
idx = faculty[faculty['name'].str.lower().str.contains(name.lower())].index[0]
display(Image(url=divs[idx].find('img').get('src'), width=200, height=200))
show_picture('marina')

Recall this HTML document.
<html>
<head>
<title>3*Global-2026-02-04</title>
</head>
<body>
<h1>Spotify Top 3 - Global</h1>
<table>
<tr class='heading'>
<th>Rank</th>
<th>Artist</th>
<th>Song</th>
</tr>
<tr class=1>
<td>1</td>
<td>Djo</td>
<td><a href='https://open.spotify.com/track/3qhlB30KknSejmIvZZLjOD'>End of Beginning</a></td>
</tr>
<tr class=2>
<td>2</td>
<td>Harry Styles</td>
<td><a href='https://open.spotify.com/track/45Z3m6yazmAi4jZuW0tzW0'>Aperture</a></td>
</tr>
<tr class=3>
<td>3</td>
<td>Taylor Swift</td>
<td><a href='https://open.spotify.com/track/53iuhJlwXhSER5J2IYYv1W'>The Fate of Ophelia</a></td>
</tr>
</table>
</body>
</html>
# Create a BeautifulSoup object from the Spotify data.
soup = bs4.BeautifulSoup((Path('data') / 'example3.html').read_text())
Part 4: Complete the implementation of the function rank_n, which takes in a positive integer n and returns the name of the n-th ranked song in the HTML document. For instance, rank_n(3) should evaluate to "The Fate of Ophelia".
Note: Your implementation should also work in the case that the page contains more than 3 songs.
def rank_n(n):
return soup.find("tr", attrs=__(a)__).find_all("td")__(b)__
soup.find and soup.find_all are the functions you will use most often.Regular expressions!